Release Notes
The release notes describe what data were released with each assay, any possible updates on cell lines, data, or software, and findings that might help users interpret the data. The release notes might also include bugs fixes or known issues and workarounds.
Data Release Note: Neurofilament Light Chain (NfL)
We are pleased to announce the release of Neurofilament Light Chain (NfL) data on Neuromine. This dataset was generated using the Siemens Atellica® Immunoassay on serum from 911 participants. The cohort comprises 95 control participants and 816 case participants.
New Files Available For Analysis
The “NfL # of Timepoints” column correlates to the number of timepoint data samples available for each participant. The full NfL data is available for analysis and can be found on the data download page under Metadata and Clinical Data. To download, click either button for the NfL folder. Please see the README documentation for more information on the full data.
New Individual Participant Data
Proteomics: 295 participant L1-L2 data has been added for a new total of 499.
Whole Genome Sequencing: 73 participants L2-L3 whole genome sequencing data have been added for a new total of 939.
New Level 4 Data
- Updated: Proteomics:
- Pre-Batch Corrected Data: New peptides, proteins, and intensities matrices for 447 released participants are available.
- Batch Corrected Data: New peptides, proteins, and intensities matrices for 436 released participants are available.
- NEW: Three File Types:
- Peptides Matrix: Maps each amino acid sequence to the corresponding peptide identified by mass spectrometry
- Protein Matrix: Maps fragments from each sample to identified peptide types.
- FC Matrix: The log 2 fold change (logarithmic scale using base 2) of each protein quantity between case and control, as well as the false discovery rate (proportion of false positives).
- NEW: Three File Types:
- Updated: Genomics: A joint genotype for 939 released participants is available
- NEW: Two File Types:
- jgt_vcf: A joint-genotyped VCF including all 939 genomes. This is a VCF joint-genotyped from the gVCF’s separated by each chromosome. Note, that previous releases did not separate data by chromosome.
- Report: A tab-delimited annotation of all of the variants separated into exonic and regulatory variants. Regulatory reports have different columns than the exonic because there is less information available for annotation.
- NEW: Two File Types:
A Special Note on Proteomics Data
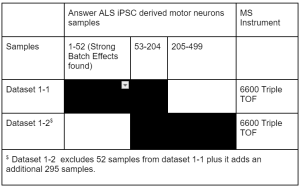
52 samples from release one have been found to have strong batch effects. The extent to which these samples vary from the subsequent data has led us to advise users not to utilize these samples in their analyses. Additionally, these samples will not be included in the level 4 proteomics data. However, we are keeping the level one and level two individual participant data available for download from the Neuromine data portal.
Level one and level two proteomics data is available on the Neuromine data portal for both datasets 1-1 and 1-2 in the table below.
Level four proteomics data is available for data set 1-2 on the Neuromine data portal. If you would like to access the proteomics level four matrix that was previously available on the portal prior to release 6.1, please contact terri@answerals.org. This matrix contains samples corresponding to dataset 1-2 in the table below.
52 Proteomic Samples with Strong Batch Effects
New Files Available For Analysis
The following files are available through Azure in the Level 3 CTRL-NEUEU392AE8 folder for each omic type. They can also be found on the main menu Data Information tab.
New Individual Participant Data
Epigenomics: 330 participant L1-L3 data has been added
Transcriptomics: 361 participant L1-L3 data has been added
New Level 4 Data
We are releasing updated level 4 matrix data for Transcriptomics, Proteomics, and Epigenomics.
- Updated: Transcriptomics: A counts matrix for 651 released participants is available.
- NEW: Epigenomics: A counts matrix for the 620 released participants. (Generated using ENCODE-DDC Pipeline Version 2.2.0)
New Additional Data
- Clinical Data
All available clinical information, including ongoing longitudinal data for applicable participants, has been updated.
- Updated – Motor Neuron Cell Line QC Staining Information
All available QC information has been updated.
Portal Updates
- Portal Layout
The data portal has received modifications in layout and design. The presentation of portal data summaries has been updated to reflect better an overview of important and relevant findings for users. Website navigation and available information have been updated.
- Data Access
To receive access to data, users will now need to complete a data access request that can be found in the portal. Each group/lab will submit one form for data access and must provide a business contact and a lead investigator. All researchers using the data must be listed in the request as collaborators. All researchers, the business official, and the lead investigator must sign a Data Use Agreement (DUA) via DocuSign before access to data will be granted.
- Data Download
Be sure to update your download tool by following the instructions on the download data page.
Data Visualization – Version Two Updates Available!
- UI Updates:
Material Design widgets were added to replace old widgets.
- Ability to switch data via an added data switcher button
Quickstart guide popup has been added at the start of the loading page. It will automatically be removed when a new plot is selected.
- New! Proteomics Tool
- New KEGG Pathways added for proteomics datasets, including:
- Alzheimers Disease
- ALS
- Neurodegeneration Pathways (Multiple Diseases)
- Thermogensis
- Salmonella Infection
- Ribosome
- Prion Disease
- Parkinson Disease
- Metabolic Disease
- Huntington Disease
- Endocytosis
- Diabetic Cardiomyopathy
- Chemical Carcinogenesis
- Oxidative Phosphorylation
- Neutrophil Extracellular Formation
- Apoptosis
- Inflammation related pathways (Toll-like receptor pathway)
- Neutrophin signaling pathway
- Endoplasmic Reticulum (ER) stress-related pathways (Protein Processing in endoplasmic reticulum)
- MAPK signaling pathway
- PI3k-AKT signaling pathway
- Plot Updates:
The following plots are now available: Dotplot, distplot, and violin plot.
New Individual Participant Data
Epigenomics: 146 participant L1-L3 data has been added
Transcriptomics: 146 participant L1-L3 data has been added
Proteomics: 72 participants L1-L2 data has been added
New Level 4 Data!
We have released level 4 data for Genomics, Transcriptomics, and Proteomics.
Genomics: Joint Genotyping of 866 released participants is now available.
Transcriptomics: a counts matrix for the 290 released participants is available.
Proteomics: an intensities matrix for 204 released participants is available.
New Data Versioning Schema
Answer ALS has introduced a new versioning schema to accommodate changes in analysis pipelines or other updates that would require a new version of data.
Example:
Version 1 data: CASE-NEUAA599TMX-5312-E_1.nodup.bam
Updated data: CASE-NEUAA599TMX-5312-E-v2-release4_1.nodup.bam
The “v2” indicates this is the 2nd version of the data.
The “release4” indicates the official release where this data was introduced.
We applied this schema in the following due to updated pipelines:
- Epigenomics – levels 2 and 3 data in releases 4 and 5. ENCODE-DCC ATAC-Seq pipeline updated from 1.1.7 to 1.7.1
- Transcriptomics – levels 2 and 3 in release 5. ENSEMBL annotation updated to version 104. All previously released samples have been re-run and labeled with “v2-release5”. All future data will be produced with the updated pipeline and therefore be considered version 1 for future releases.
- Genomics – level 3 ExpansionHunter VCF in release 3. ExpansionHunter pipeline standardized to version 2.5.5
With each release, aggregate data products (level 4) may be updated to include the newly released samples. The following example depicts the number of participants in this aggregate file (n=290) and applies the new versioning schema.
Example:
AnswerALS-290-T-v1-release5_rna-counts-matrix.csv
Portal Updates
Portal Index and Datatable Updates
- PLS Column: added a subgroup for Primary Lateral Sclerosis in Non-ALS MND participants in the portal datatable and faceted search
- iPSC/diMN Inventory Updates: updated the iPSC and diMN inventory from Cedars in the portal datatable and search
- WGS Variants Updates: updated the variants data from the June analysis performed by Dr. Finkbeiner’s group at the Gladstone Institutes.
Assay and Pipeline Descriptions
A new page with information on sample preparation, library construction, and pipeline methods has been added to the main menu of the data portal site.
Card View Update
Cards on the Data Search webpage can be moved by the user via drag and drop to provide a customized view.
Table Update
Columns can be managed by users. Add, delete, or reorder columns for a personalized view.
CLI Download Progress Update
Stabilized the display of download progress in the CLI. Progress output is now displayed in summary in the CLI and large volumes of progress outputs are directed to the user’s log file.
Multiple Emails for a single user
Reconciled an issue to consolidate multiple accounts for the same user into one account. Previously, users could create more than one account using different sign-in methods.
Azure to Azure transfers using the Download Tool
The download tool now supports downloads within Azure from Answer ALS stores to user stores in the same region, reducing costs to the program and to users ’ allotted download credits.
Release Package
We have released Proteomic data (wiff, wiff.scan, mzML, and counts matrix) for 136 participants to complement the existing 150 transcriptomic (FASTQ, BAM) and epigenomic (FASTQ, BAM, Peaks) releases. This proteomic release overlaps with 126 of our previously released participant wgs data.
Full release metadata Package:
- We’ve included complete clinical metadata, participant and inventory metadata and the Portal datatable
- All metadata tables are keyed by “Pariticipant ID” (CASE/CONTROL + GUID) to allow for easy joining of tables
Portal updates:
- Users are now alerted to the number of participants associated with each data type in the data download page
- The download tool command-line-interface (CLI) accepts relative paths and has improved help instructions
Proteomic data descriptions:
The DIA data analysis was performed (on the profile .mzML file format) using openSWATH running on an internal high-performance computing cluster at Cedars-Sinai Medical Center. The Mass Spec data was only searched against the human database, therefore human is implied in the Level 4 data counts.
- Level 1: .wiff and .wiff.scan files are raw per-sample data generated by the Sciex TTOF 6600 instrument
- Level 2: .mzML profile files are per sample files obtained by converting the wiff files using ProteoWizard (v.3.0.6002). For downstream processing, we need to convert this vendor-format (.wiff) to an open-source format (.mzML).
- Level 3: Answer ALS proteomics data does not contain a more processed per-sample level 3 data type
- Level 4: one .csv file contains the counts data for all proteomics samples in this release for a specified uniprotID and protein name. Please note that a zero (0) in this data indicates the protein was not detected. While data levels 1-3 include single-sample data that are not expected to change over time, the Level 4 data will be updated to reflect the samples included in each release.
NOTE: For this release, there is a known display issue when viewing the calculation for level 4 proteomic data. The size of the level 4 download file appears to be 700MB while the actual download size is ~5MB. This display issue will not affect your download and is expected to be resolved in the next portal update (February). Status update: This display issue has been resolved (February 2021)
Release 4.0 contains new epigenomic and transcriptomic data for an additional 80 participants bringing the total data released for these two assays to 150 total participants. We are also planning to release the corresponding participant proteomic data within the next 4 weeks.
This data release coincides with the unveiling of the new Answer ALS data portal. We hope the many changes to visualizing our metadata, data, and download options will improve user experience.
We have released an additional set of Whole-Genome Sequencing (WGS) data, levels 2-3 (CRAM and VCF), for 475 Participants.
This brings the total number of participants with WGS data to 866!
NOTE: Level 1 data for WGS (raw FASTQs) have been moved from general access and are available by special request.
Please contact us if you require this data.
Data Replacement: WGS ExpansionHunter VCFs (files ending in “eh.vcf”) have been re-run and standardized using Illumina’s Version 2.5.5 software.
The data portal has a new landing page and release notes tab.
We have added the following Transcriptomics data:
- 86 Indexed BAM files
- 86 Counts files
We have now released 391 WGS Samples, 85 Epigenomics samples (FASTQ, BAM, Peaks), and 86 Transcriptomics samples (FASTQ, BAM).
Full release metadata package:
- We’ve added complete Clinical metadata, Participant/Inventory metadata and Portal metadata.
- All metadata tables are keyed by “Participant_ID” (CASE/CONTROL + GUID) to allow for easy joining of tables.
We have removed NeuroLINCS data and the “Analyze” tab that previously used this data.
Users are now alerted to the size of data within the download script.
Sex Effects in RNA-Seq data:
- The standard step after obtaining level 3 data (Counts) is statistical inference of systematic changes between conditions (e.g ALS and CTR) by modeling gene expression data with a binary variable with two levels (design ~condition, condition =0 for CTR, 1 for ALS). In the presence of confounding factors a more complex design (e.g. design~batch + condition) may be needed to exact the disease relevant signal while controlling for the confounders.
- In this data release, one of the dominating contributors to the gene expression changes observed in our initial principal component (PCA) analysis is the inherent sex effect. Genes that contribute to the sex effect include both sex chromosome linked genes and autosomal genes. This gender specific gene expression has also been reported in post mortem human brains, as well as IPSCs. In order to extract disease relevant signal, we recommend controlling sex effect for your downstream analysis. For example, excluding chrX, chrY linked genes and adding an additional binary variable to the design account for sex effect.