Data Information
Answer ALS is dedicated to building the largest ALS data set, both clinical and OMIC data, and sharing it with researchers. We want to make the data easy to use and understandable for our users. Below are descriptions of the various data we collect and examples of how scientists use the data in their research on ALS.
Answer ALS
The clinical data tables and files included in the download package are essential resources for researchers seeking to gain insights. These tables and files contain detailed information about patient demographics and medical histories. Researchers can use this data to conduct analyses, generate statistics, and develop models to better understand disease progression and treatment effectiveness.
What’s included in the package:
aals_dataportal_datatable.csv – A table view of high-level summary information keyed by participant ID. Information includes demographics, cell line QC and availability.
aals_participants v2.csv – A list of participants by ID indicating cohort group.
aals_released_files.csv – A list of released files with the data path and size of each indicated.
Clinical_Dictionary_v2_2023.csv – A reformatted and curated clinical dictionary for easier analysis. Download the file here.
Clinical_Dictionary.csv – Raw data dictionary.
Clinical Folder – This folder holds 33 csv files containing clinical measurements from de-identified participant visits.
ALS TDI
ALS TDI has partnered with Answer ALS to put clinical and OMICs data from the ARC Study on the Neuromine Data Portal for researchers around the world to use. Data from the ARC Study will be updated on a yearly basis. Data for U.S. based ARC participants that have contributed OMICs data are shared on Neuromine. For access to the rest of the ARC Study data, you can visit the page here and fill out a contact form
The clinical data tables and files included in the download package are essential resources for researchers seeking to gain insights. These tables and files contain detailed information about patient demographics and medical histories. Researchers can use this data to conduct analyses, generate statistics, and develop models to better understand disease progression and treatment effectiveness.
What’s included in the package:
alstdiarc_dataportal_datatable.csv – A table view of high-level summary information keyed by participant ID. Information includes demographics, cell line QC and availability.
alstdiarc_participants.csv – A list of participants by ID indicating cohort group.
alstdiarc_released_files.csv – A list of released files with the data path and size of each indicated.
Clinical_Dictionary_2025.csv – A reformatted and curated clinical dictionary for easier analysis. Download the file here.
Clinical_Dictionary.csv – Raw data dictionary.
Clinical Folder – This folder holds 33 csv files containing clinical measurements from de-identified participant visits.
Level 1
Level 2
Level 3
Level 4
Level 5
Level 1
Level 1 data is raw, immutable data coming off an instrument (e.g. a sequencer). For genomics, we only provide data corresponding to this level upon special request.
Level 2
Level 2 data is raw data mapped against the appropriate reference. Genomics (wgs) uses hg38 as a reference.
Level 3
Level 3 data the most processed form of patient-specific data.
Level 4
Level 4 data is attained from the joining of a cohort of patients’ level 3 data from a particular assay.
Level 5
Level 5 data is comparitive data between two specific cohorts typically labelled as differentially expressed genes (DESeq) or proteins or enriched sites within the genome (DiffBind). Data coming soon!
Welcome to the Assays and Pipelines Information Hub! Here, you will find a comprehensive collection of information on the various assays and pipelines used in the generation of the Answer ALS and ALS TDI data. Our goal is to provide you with a detailed overview of the techniques and tools used in the data generation process, including their purpose, methodologies, and applications. Whether you are a researcher or computational scientist, you will find this resource to be an invaluable tool for understanding the complex world of data generation. Explore and learn more about the assays and pipelines used by the ALS TDI and Answer ALS researchers!
Follow this link for detailed information.
This page is dedicated to showcasing real-world examples of our data in action. Our aim is to provide you with some data-use examples. By exploring these examples, you will gain a better understanding of the power and versatility of our data and how it is being used to drive innovation and solve complex problems. Whether you are a student, researcher, data scientist, or simply curious, this resource is an excellent starting point for learning about the exciting world of data. So, dive in and start exploring the data-use examples that we have curated for you!
Our Release Notes page has all the information you need about our datasets’ latest updates and releases. This page is designed to provide you with a summary of what has changed in each release of our datasets, and includes any new features, improvements, and bug fixes available for the data portal site. Our release notes allow you to stay up-to-date with the latest data and ensure that your work remains accurate and relevant. Bookmark this page and check it regularly to stay ahead of the curve and make the most of the data that we have to offer.
The use of a sample mapping file in controlling pipeline analysis for batch effects plays a crucial role in ensuring the reliability and reproducibility of results, especially in complex biological experiments like cell line differentiation studies. Batch effects can significantly skew data interpretation because they introduce variation that is not related to the biological variables being studied. Use the Sample Mapping file with the readme text file to understand how to incorporate this into your analysis of the Answer ALS data.
The sample mapping information are available in the Level 3 folder for each omic type. They can also be found on the main menu Resources tab.
A Special Note on Proteomics Data
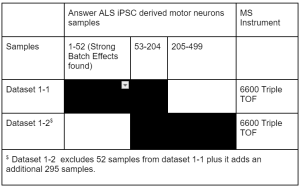
52 samples from release one have been found to have strong batch effects. The extent to which these samples vary from the subsequent data has led us to advise users not to utilize these samples in their analyses. Additionally, these samples will not be included in the level 4 proteomics data. However, we are keeping the level one and level two individual participant data available for download from the Neuromine data portal.
Level one and level two proteomics data is available on the Neuromine data portal for both datasets 1-1 and 1-2 in the table below.
Level four proteomics data is available for data set 1-2 on the Neuromine data portal. If you would like to access the proteomics level four matrix that was previously available on the portal prior to release 6.1, please contact terri@answerals.org. This matrix contains samples corresponding to dataset 1-2 in the table below.